DeepMind AI makes a science-shaking protein breakthrough
Alphabet's DeepMind has used its AI system to figure out a five decade old mystery in biology, the company has announced today, using AlphaFold to help understand protein behaviors. The company came to notoriety for its neural network developments, which demonstrated their human-besting capabilities in playing chess, go, and shogi.
Google acquired DeepMind in 2014 – not without a little controversy along the way – and it became a subsidiary of Alphabet in 2015. AlphaGo, its go AI, beat the human world champion player the following year, while AlphaZero went on to show how reinforcement learning could be used to effectively train the AI through play against itself.
AlphaFold, though, tackles a very different challenge. The "protein folding problem" is shorthand for attempts to understand how the amino acid sequence in a protein shapes its 3D atomic structure. That form is led by the underlying folding code which takes into account thermodynamics and interatomic forces; the protein structure prediction of trying to understand a protein's native structure from the animo acid sequence; and the kinetics of how the fold itself takes place.
While it sounds esoteric, understanding how amino acids operate is believed to be key to a number of challenges in biology. That includes everything from addressing diseases in humans, through to broader applications such as enzymes that break down plastics or other waste.
The goal has been to come up with a computational method to predict folding, rather than an experimental one, which could be faster and more efficiency. "A major challenge, however, is that the number of ways a protein could theoretically fold before settling into its final 3D structure is astronomical," DeepMind points out.
A challenge was set up in 1994, CASP, to pit predictive methods against each other in the hunt for a computational solution. Its measure of success is the so-called Global Distance Test, or GDT, which is based on the percentage of animo acid residues that are predicted within a threshold distance of their correct position. It's scored from 0-100, with the unofficial benchmark being anything over 90 GDT as comparable with experimental findings.
Today, DeepMind says, its attempt in the fourteenth challenge – CASP14 – scored 92.4 GDT. "This means that our predictions have an average error (RMSD) of approximately 1.6 Angstroms," the company says, "which is comparable to the width of an atom (or 0.1 of a nanometer)."
It's a significant jump over DeepMind's 2018 entry – the last CASP to run – which saw the previous AlphaFold generation fail to reach 60 GDT.
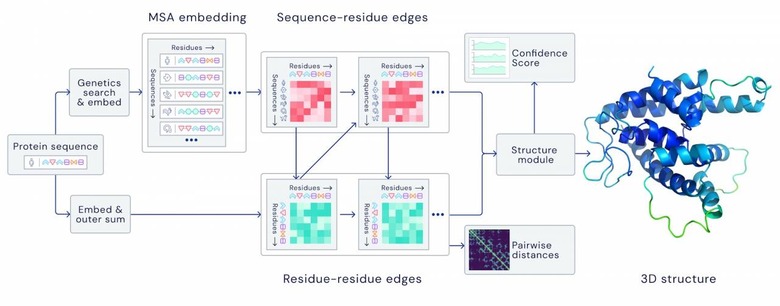
"For the latest version of AlphaFold, used at CASP14, we created an attention-based neural network system, trained end-to-end, that attempts to interpret the structure of this graph, while reasoning over the implicit graph that it's building," DeepMind explains. "It uses evolutionarily related sequences, multiple sequence alignment (MSA), and a representation of amino acid residue pairs to refine this graph."
DeepMind uses the latest generation of Google's TPU neural processing cores – approximately 128 of them – with around 170,000 protein structures from public databases together with other protein sequence databases. It took "a few weeks" to crunch, the company says. Next up, the hope is to make access to the system in a scalable way for third-party researchers, while applying the technology to better understand how protein structures impact specific diseases and could shape drug development.
